 |
|
The Flask
|
This exercise demonstrates how a problem can be mapped onto the Flask model and solved by running the Flask simulator. Figure 1 shows a graph with 8 nodes labeled “a” through “h”. The arcs between the nodes represent a distance. The problem is to discover the shortest distance between “a” and “g”.
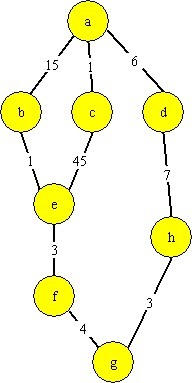
Figure 1 - The cities and the distances between them by various roads
There are 3 paths from “a” to “g”: <a,b,e,f,g>,
<a,c,e,f,g>, and
<a,d,h,g>. The latter is the
shortest.
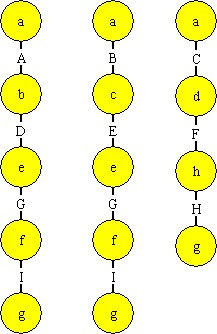
Figure 2 - Complete molecules that represent paths from city a to city g
Mechanisms are also needed to flag a molecule as complete when it contains all the nodes on a path from “a” to “g” and a way to select a molecule that represents a good solution. For the former, we create a class of atom labeled “S”. It has two and binding sites “S” and “E” (for start and end) and a conditional binding site labeled “XX”. “S” and “E” are electrostatic. We add an “S” binding site to the “a” atoms and an “EE” binding site to the “g” atoms. When the “S” atom attaches to an “a” and a “g” atom, it then exposes its “X” binding site. This site exposes the molecule as being a solution to the path length problem.
The second mechanism needed to select the best solution molecule is provided by the Improved Bond Length Comparator (ImprovedBLC) atomic machine, which is labeled “Comparator” in this example. Unlike the other atoms, which have multiple copies, there is only one “Comparator” atom. Its job is to attach to two molecules at a time and compare their path lengths. The molecule with the shortest path remains attached to the comparator. The second molecule is released after its “X” binding site is removed. This prevents the molecule with the less optimal solution from being selected for comparison again. If two molecules have the same path length, one is detached with its “X” binding site removed.

Figure 3 - Atoms for the path length problem
Note that it is possible for partial solutions to
form that have an
exposed "X" binding site since an an "S" atom could bind to an "a" and
a "g" atom before the full path is formed. Due to the relative
concentrations of the atoms, it is unlikely that a partially formed
solution will bind to the Comparator. However, a better solution
can be modeled after the second Traveling Salesman
Problem using IntraMolecular binding
sites.
To set up the problem, the atoms described
diagrammatically
in Figure 3 must be generated in the Flask in concentrations high
enough so
that molecules of all the path solutions are formed.
A good starting point is to have all the atoms except the
comparator have number 10. There is only
one comparator. The comparator atom has
three booleans that
can be set from the FlaskRunner GUI or from program calls:
doMax, print,
and dissolve.
The doMax boolean
indicates that the comparator is looking for maximum path lengths. In this problem, we want to find the shortest
path, so doMax should be
false. The
print parameter indicates
that the atomic machine should print to
standard output
each time it makes a comparison. Print
should be false when running from the GUI.
Finally, dissolve
indicates that the molecule that loses the
comparison
should be prevented from bonding to the comparator again by removing
the
binding site via which it connects to the comparator.
Dissolve should be true.
When run from FlaskRunner, the View Molecules features displays the molecules created by the simulation run. Clicking on the comparator atom will cause FlaskRunner to create a dialog window with the minimum path length found. Figure 4 shows a sample solution using the MoleViewer feature of the FlaskRunner application.
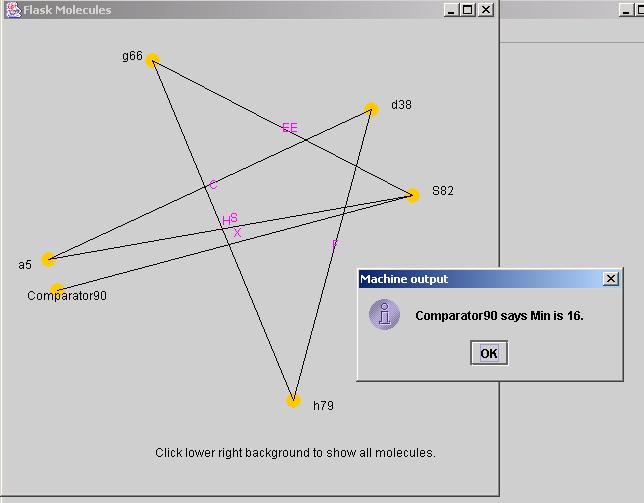
Figure 4 - MoleView of one solution

